Investigating the Genetic Basis of SURFS and MAS Using an Exome Slice Approach
Michael Matt, MD; Wenying Zhang MD, PhD, MBA, FACMG; Elizabeth Baker; Alexei A. Grom, MD; Rebecca Marsh, MD; Grant Schulert, MD, PhD

In this project we sought to better understand the genetic underpinnings of systemic autoinflammatory syndromes. Our exome slice data revealed likely diagnostic genetic variants in 8/33 patients, many of which are located in genes implicated in familial HLH markers to evaluate the best ways to nutritionally support these infants through WIC.
Michael Matt, MD
Abstract
Background: Children with autoinflammatory disorders including systemic undefined recurrent fever syndrome (SURFS) and macrophage activation syndrome (MAS) often lack a genetic diagnosis.
Objective: We sought to define the utility of an “exome slice” approach to examine the genetic basis of patients with SURFS and/or MAS.
Methods: 37 patients less than 18 years of age with the diagnoses listed above were enrolled in this IRB-approved study and genetic samples were obtained after an informed consent process. Targeted exome sequencing was performed for 394 genes implicated in known inflammatory disorders as well as primary immunodeficiency syndromes. Variants within exons and the flanking sequences were evaluated using an in-house bioinformatics pipeline including GATK3.5 and Alamut Batch 1.4.4 software.
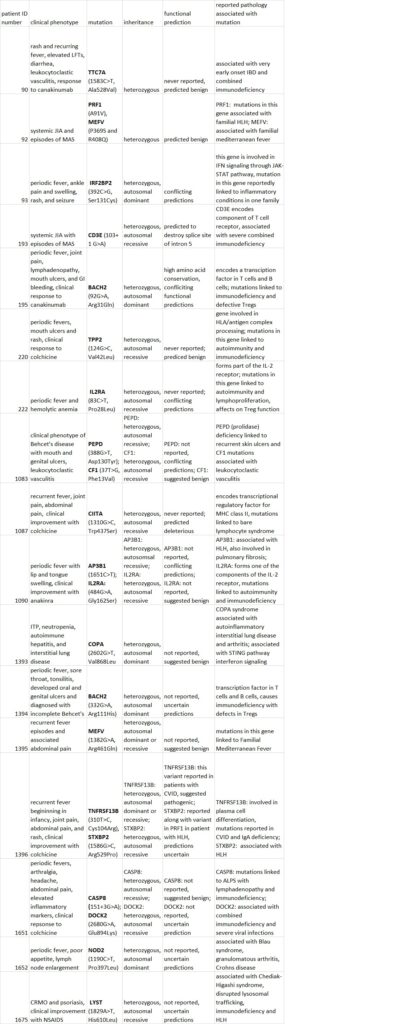
Results: This exome slice approach detected likely diagnostic variants in 8/33 patients with SURFS and/or MAS. We assessed these variants as likely diagnostic based on functional predictions or significant overlap between the patient’s clinical phenotype and reported cases in the literature (Figure 1). Variants in this cohort involved known monogenic causes of periodic fever syndromes (MEFV, MVK), genes implicated in cases of familial HLH (PRF1, STX11, and STXBP2), and mediators of interferon signaling (COPA, STING1). We also identified and characterized a list of variants of uncertain significance which were never reported in the Human Gene Mutation Database (HGMD) or had conflicting in silico functional predictions (Figure 2). Variants in this cohort again involved known autoinflammatory genes (MEFV (2) and NOD2), as well as genes associated with familial HLH (PRF1, STXBP2, and AP3B1). Notably, however, numerous patients in this cohort carried variants in genes implicated in autoimmunity and immunodeficiency (BACH2 (2), IL2RA (2), CD3E, TPP2, CIITA, TNFRSF13B, DOCK2). Finally, as validation, we found that this approach identified and confirmed known mutations (MVK (2), MEFV, and STING1) in four patients with defined autoinflammatory disorders.
Conclusions: This report demonstrates the ability of an “exome slice” approach to identify potentially significant genetic variants in a cohort of children with SURFS and/or MAS. The genetic landscape of these autoinflammatory patients includes genetic variants in known periodic fever genes, genes associated with familial HLH, and variants involved with immunodeficiency.
Figure 1
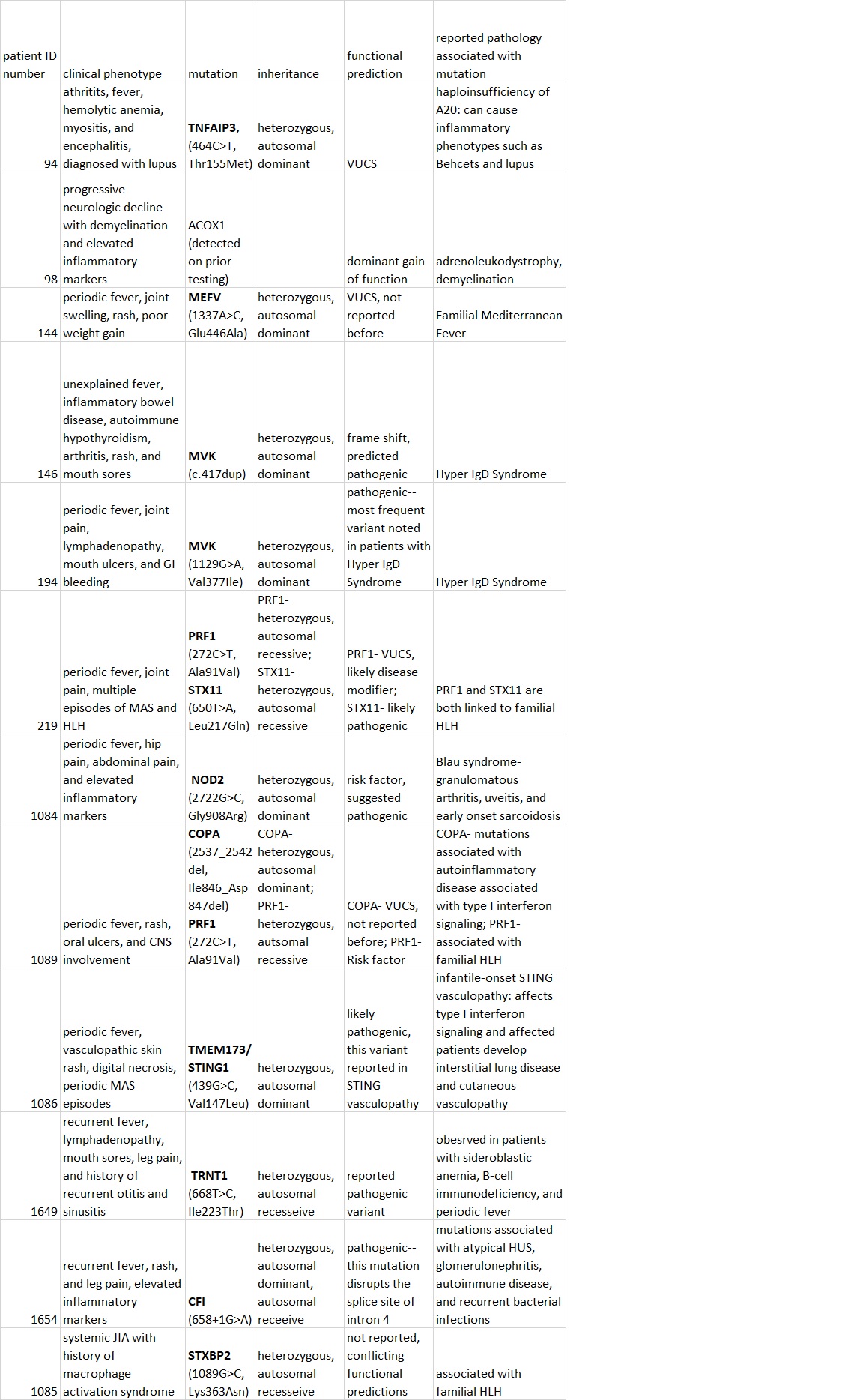
Figure 2